David Jacques
Understanding the Interactions that Enable Evasion
David Jacques is teasing apart the molecular interactions between viral and host proteins to figure out how viruses like HIV escape cellular defence networks.
Once viruses get into host cells, they need to run a gauntlet of molecular sensors and antiviral systems before they can replicate and spread to the next cell. Viruses have many ways of escaping detection: sometimes they outrun the defences, sometimes they hide, and sometimes they fight back.
David believes that one of the best ways to understand how viruses evade host defences—and discover new ways to stop viruses in their tracks—is to investigate the structures of the host and viral proteins while they are coupled together, using protein crystallography, or with cryo-electron microscopy.
Viruses exist because they have managed to find ways to subvert and control normal cellular functions. They achieve this through exquisitely specific molecular interactions with their hosts. David is interested in understanding the nature of these interactions. Understanding of host-virus interactions not only provides scope for the development of novel therapeutic antivirals, but often grants key insight into the molecular functions and defences of the host. Some examples of fundamental discoveries made through virus research include the discovery of oncogenes, restriction enzymes, and RNA sensing. David is particularly interested understanding the mechanisms by which viruses escape innate immune detection, and has a particular focus on HIV – a virus that specifically infects cells of the immune system.
About David Jacques (he/him)
Dr David Jacques leads the Structural Virology Group at the EMBL Australia Node for Single Molecule Science. Having completed a Bachelor of Science, majoring in biochemistry and chemistry, David undertook his PhD under the supervision of Professors Jill Trewhella and Mitchell Guss at the University of Sydney. During that time, David employed both crystallography and small-angle scattering to understand protein structure. He also authored the internationally accepted standards by which biological small-angle scattering data are now reported in publications. In 2012, the award of an NHMRC Early Career Fellowship enabled him to join the group of Leo James at the MRC Laboratory of Molecular Biology, Cambridge. Here, he studied the role played by the HIV capsid during the early stage of infection, revealing structural dynamics and interactions with the host at a molecular level not previously seen. Since returning to Australia, David leads an independent research team focused on understanding the molecular details of host-pathogen interactions through the use of crystallographic, single molecule fluorescence, and cryo-electron microscopy methods.
Select Publications
More Information
In the News
- How does HIV get into the cell's centre to kick start infection?
- How HIV adapted for pandemic spread
- UNSW researchers finalists in 2019 ‘Oscars’ of Australian science
- Science without sight: bringing medical discovery to low vision community
Career Highlights
- 2016-2018 NHMRC Early Career Fellowship
- 2014-2016 MRC Careeer Development Fellowship
- 2014 Paper of the Year award from Journal of Biological Chemistry
- 2012 Young Investigator Prize, Lorne Conference on Protein Structure and Function
- 2006 Thompson Prize, Sydney Protein Group
Projects
Regulation of DNA Synthesis and Host Evasion by Lentivirus Capsids
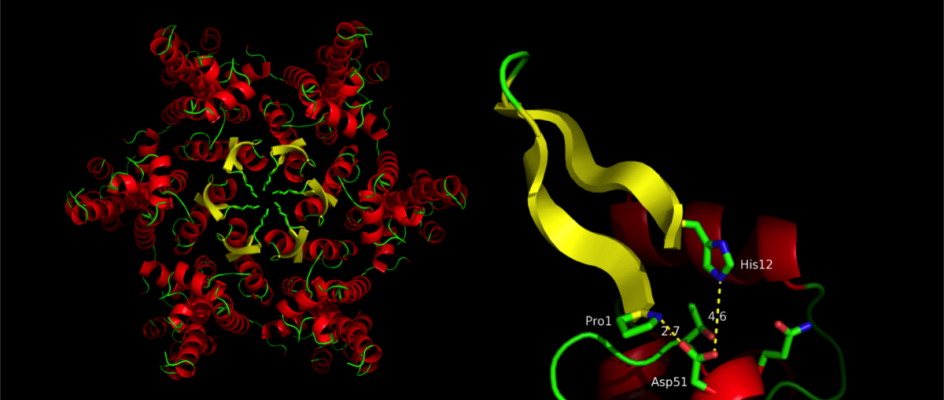
 The retroviruses are a family of viruses that carry their genetic material on two copies of a single-stranded RNA genome within a protein shell called a capsid. During infection, reverse transcriptase converts the RNA genome into double stranded DNA. This newly synthesised DNA is then integrated into a chromosome within the host cell, thereby establishing infection. The lentiviruses are a genus within the retrovirus family that have the additional ability to infect non-dividing cells, meaning that they are capable of crossing the nuclear envelope. This property makes lentiviruses insidious pathogens as they can infect the non-dividing cells of the immune system and cannot be cleared without the destruction of these cells. HIV is an example of such a virus. Surprisingly, it is the capsid that gives lentiviruses the ability to cross the nuclear envelope. Recently, we have shown that the capsid determines whether or not viral DNA is detected by host pattern recognition receptors. This project aims to determine how these capsids engage with host proteins to regulate DNA synthesis, while simultaneously avoiding innate immune detection.
The retroviruses are a family of viruses that carry their genetic material on two copies of a single-stranded RNA genome within a protein shell called a capsid. During infection, reverse transcriptase converts the RNA genome into double stranded DNA. This newly synthesised DNA is then integrated into a chromosome within the host cell, thereby establishing infection. The lentiviruses are a genus within the retrovirus family that have the additional ability to infect non-dividing cells, meaning that they are capable of crossing the nuclear envelope. This property makes lentiviruses insidious pathogens as they can infect the non-dividing cells of the immune system and cannot be cleared without the destruction of these cells. HIV is an example of such a virus. Surprisingly, it is the capsid that gives lentiviruses the ability to cross the nuclear envelope. Recently, we have shown that the capsid determines whether or not viral DNA is detected by host pattern recognition receptors. This project aims to determine how these capsids engage with host proteins to regulate DNA synthesis, while simultaneously avoiding innate immune detection.
Understanding the Role of Viral Accessory Proteins
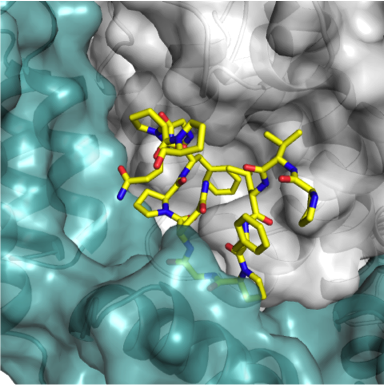
Most human viruses carry genes that code for proteins that are not necessarily packed into the viral particle. These accessory proteins frequently play roles in hijacking host cell processes and evading host immunity. While some of these proteins are well understood (for example Vif and Vpu from HIV which antagonise the host defense proteins APOBEC3G and Tethering, respectively), there remain many that are poorly characterised. The difficulty in determining roles for these proteins is three-fold. Firstly, they are poorly structured until they recognize their target, making recombinant production and structure determination difficult. Secondly, they may have multiple targets. And thirdly, their targets may be involved in multiple processes, leading to complex cellular phenotypes. To address this, we will be using a novel ‘bottom-up’ approach, which screens interactions using single molecule fluorescence. Our novel screening approach aims to determine functional roles for a subset of proteins from diverse virus families, including retro-, pox, and paramyxoviruses.
Collaborators
- Dr Leo James, MRC Laboratory of Molecular Biology
- Prof Greg Towers, University College London
- Prof Till Boecking, UNSW
- A/Prof Yann Gambin, UNSW
- A/Prof Emma Sierecki, UNSW
Current Funding Sources
- Wellcome Trust
- Australian Research Council
- National Health and Medical Research Council
- UNSW Sydney