Electrical and chemical signals generated within cells, tissues and organ systems drive vital functions. Izzy Jayasinghe and her team investigate how these signals are relayed to trigger a heartbeat, and drive other bodily functions, by combing super-resolution microscopy tools they develop with existing technologies.
Most cells contain specialised hubs of signalling proteins and accessory molecules that propagate fast, large, and repeatable signals pivotal to healthy heart rhythm and other critical functions. Now that we know these hubs are wired differently in heart failure, chronic pain, and muscle weakness, Izzy’s team will build biochemical maps of these signals to unravel disease processes, and to design and refine novel precision therapeutics.
Learn more about the Signalling Nanodomains Lab
Rapid mobilization of ions or small molecules inside cells are amongst some of the fastest signalling mechanisms fundamental to life. They underpin physiologies such as the heartbeat, muscle contraction, neurotransmitter release, activation of gene transcription, and post-translational modifications. These mechanisms are also responsible for major human diseases and disabilities such as heart disease, cancer, paralysis, and chronic pain. Many such morbidities have no cure, whilst effectiveness of pharmacological treatments tends to be variable. At the core of some of the underpinning fast signals, are intracellular calcium signals (calcium sparks, puffs, waves, or transients) in excitable cells like muscle and neural tissues. They consist of specialised, nanoscale signalling domains that harbour ion channels and accessory molecules that coordinate to generate calcium sparks.
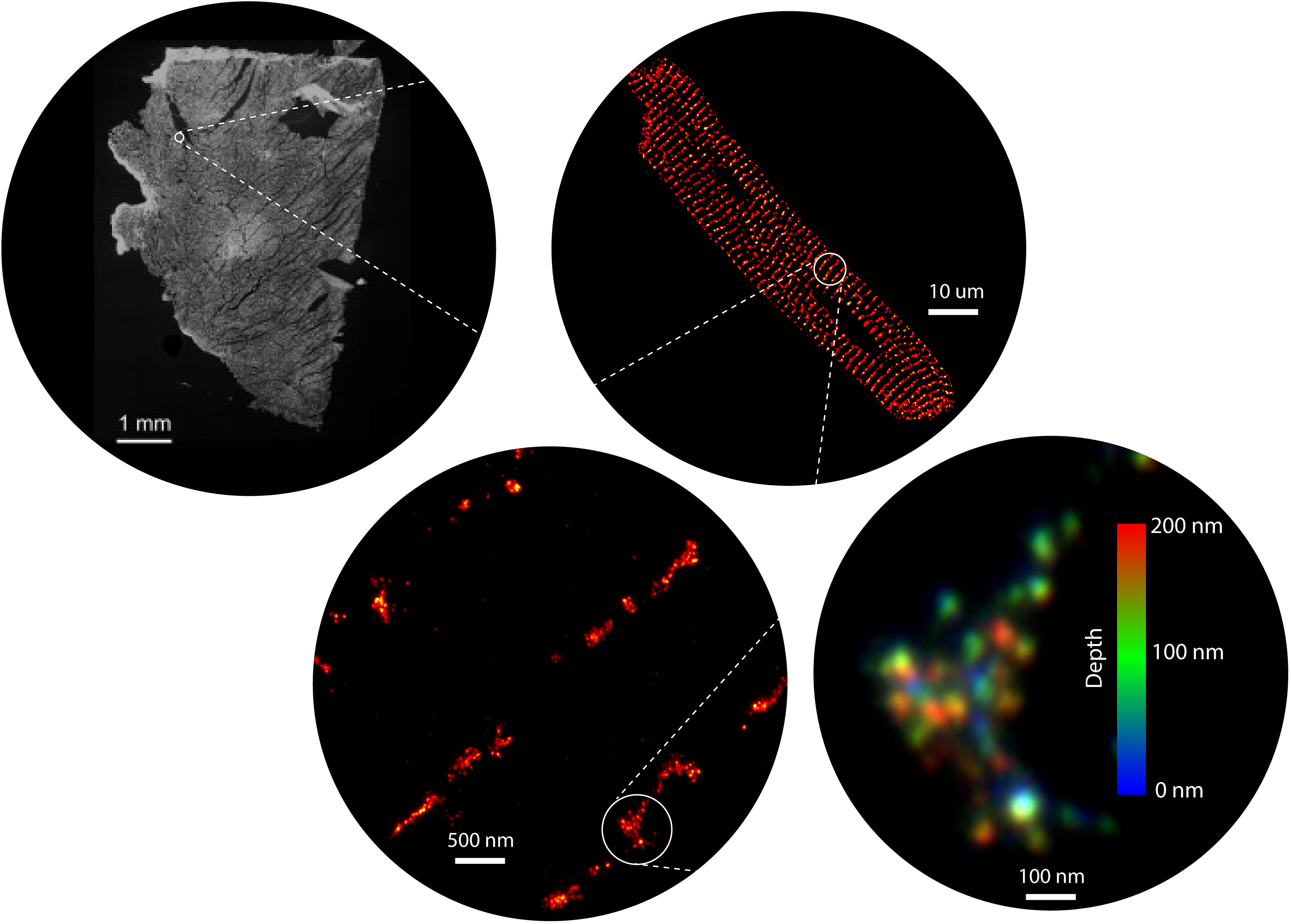
For over 15 years, our group have harnessed the power of a range of super-resolution microscopy technologies to resolve and visualise the shapes, locations, and molecular components intrinsic to these nanodomains (Jayasinghe et al, 2018). We now have the microscopy and analysis tools to not only map the position of each ion channel within these domains, but also to detect specific biochemical signatures on each channel, in situ (Sheard et al, 2019). For decades, the nanodomains and their signals have been imaged in isolation. However, one of our recent innovations, a correlative microscopy protocol (Hurley et al, 2021; Hurley et al, 2023), has allowed us to visualise the communications of channels such as ryanodine receptors can produce unexpected patterns of calcium signals. In particular, our discoveries have unearthed spatial heterogeneities in the organisation of calcium handling proteins of the myocardium. Our early observations suggest that these heterogeneities are not limited calcium handling proteins (such as ryanodine receptors (RyR2), L-type calcium channels, SERCA2A, and sodium calcium exchanger) but extend to regulatory molecules, second messengers, nucleic acids, and other structural proteins across the multitude of cells in the healthy heart. We also observe that these variabilities are accentuated in heart disease and may explain the limited effectiveness of the past and present generations of pharmacological therapies targeting the myocardial contractility..
Super-resolution microscopy, since its inception over two decades ago, has unlocked numerous secrets of the life processes. We have been one of the earliest adopters of this technology in its wide-ranging incarnations known commonly by numerous acronyms such as STORM, PALM, PAINT, STED and SIM. In spite of the nanometre-scale resolution that is on offer, access and usability of these methods remain modest due to the specialist nature of both the probes and the microscopy instrumentation. Over the past few years, our team have developed expertise in expansion microscopy (ExM) which parallels, and often exceeds, the resolution afforded through the traditional, localization- or optics-based super-resolution techniques. ExM uses molecular crosslinking and tissue clearing chemistries to obtain a three-dimensional imprint of cells, tissues, and/or whole organisms onto a polyacrylamide hydrogel (Sheard et al. 2019). These gels can then be osmotically swollen by a factor of >1000, effectively magnifying the intricate details of the sample that were previously too small to be resolved. ExM samples therefore allow us to visualise nanoscale features of cells and tissues conveniently with standard, and sometimes homebuilt, microscopes. Alongside of this methodology, we have been developing a series of tools that include high-throughput arrays, a new palette of fluorescent counterstains, distortion detection tools, gel automation robotics and 3D printable microscopy platforms to allow non-specialist microscopists to adopt super-resolution.