Dynamics of how pathogenic bacteria punch through the membranes of host cells revealed with single-molecule microscopy.
Using a new single-molecule technique developed at UNSW Sydney, Australian researchers have gained molecular insight into how bacterial proteins breach the membranes of mammalian cells.
Organisms from all domains of life are armed with specialised sets of hole-punching proteins used to attack the cells of other organisms. Mammalian immune cells use pore-forming proteins to destroy cancerous or infected cells. Bacteria, and other pathogens, use these proteins to lyse or gain entry into host cells. But researchers have not had a clear idea of exactly how individual protein molecules assemble together to form these pores, until now.
Researchers from UNSW Medicine & Health’s EMBL Australia Node in Single Molecule Science have developed a single-molecule microscopy technique to investigate this process. Together with collaborators from Monash and Melbourne Universities, they tracked the pathway of pore-formation by a bacterial protein that targets mammalian cells, called perfringolysin O. The molecular details of how this protein complex assembles itself onto the surface of plasma membranes right through to when it opens up the pores is now published in the journal eLife.
“Our single-molecule microscopy method allows us to track the assembly of individual pores. So, we were able to identify crucial steps of this pathway to find points of weakness and potential avenues for intervention” said UNSW medical researcher Associate Professor Till Böcking, who led the research team.
Identifying the pivotal steps in this assembly pathway will allow scientists to design new strategies to target specific pore-forming stages with the aim of modulating their activity in immunity and infection.
Single-molecule resolution
Using purified proteins, and liposomes as a simple model system for the target membrane, the researchers could see when the pores were formed. By loading the liposomes with a fluorescent dye, A/Prof Böcking and his team could pin-point the exact time that a pore is opened, and the dye is released.
Their experiments revealed that once two molecules bind together on the membrane, this stable interaction triggers the process of adding more pore-forming proteins to complete the ring. They also observed that pores could open even before construction of the rings were completed; an arc formed, with as few as four molecules, could create a pore opening.
“The take home message is that these are very efficient hole-punching machines that have evolved to form pores in all sorts of circumstances. We hope that understanding that the conditions actually dictate the assembly pathway gets people to rethink the way they do their experiments,” A/Prof Böcking said.
From their findings, the researchers hypothesise that the mechanism they observed with perfringolysin O could be similar for other members of this family of pore-forming proteins – called cholesterol-dependent cytolysins – but that has yet to be tested.
A/Prof Böcking believes that this new single-molecule technique could provide important insight into how other pore-forming proteins are deployed.
“It is a powerful method because it has the resolution and sensitivity to track this entire process in real-time. With this approach, we can visualise the steps and dynamics of it, which is difficult to do with other methods,” he said.
The experimental work was co-led by Conall Mc Guinness and James Walsh.
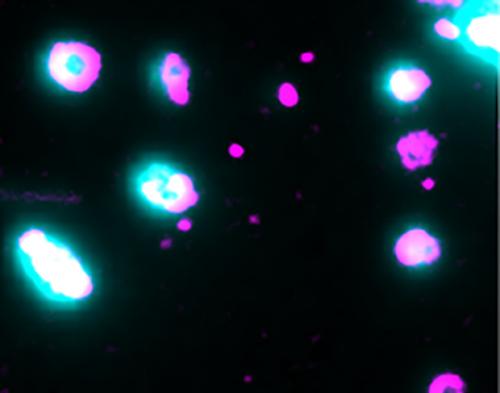
Liposomes are loaded with dye (cyan) to pin-point opening of the pores by membrane-bound pore-forming proteins (magenta). Single-molecule microscopy captures the moment the pores open and the dye is released