Inner workings of RNA concentration homeostasis in mammalian cells revealed with image-based genome-wide screening.
Mammalian cells have intricate mechanisms in place to ensure normal function and development, but many remain a mystery to scientists. An international research team have now uncovered new insights into how cells coordinate cellular amounts of messenger RNA (or mRNA) – the instructions for protein synthesis that is transcribed from DNA genetic blueprints – with their growth and size.
A study – co-led by Dr Scott Berry of UNSW Medicine & Health’s EMBL Australia Node in Single Molecule Science and University of Zurich’s Professor Lucas Pelkmans – dissects the processes at play inside mammalian cells to keep global mRNA concentrations in check.
They report in the current issue of Cell Systems that mRNA concentrations remained the same in human cell lines regardless of their size and growth cycle phase, or even after they experimentally interfered with cellular machinery responsible for the generation and breakdown of RNA.
Dr Berry explains that previous studies in yeast showed that mRNA production rates and degradation rates are “buffered”, so that when one is perturbed, the other changes to compensate. But how this relates to cell-size-scaling of RNA transcription is still unclear.
By imaging millions of cells, and extracting information from each one of them, the researchers uncovered key pathways in maintaining consistent cellular concentrations of RNA. This extensive screen consisted of 21,000 experimental conditions in which cells were treated with silencing-RNA to disable different genes associated with both RNA synthesis and degradation.
“We saw that RNA levels scaled with cell size. The bigger the cells were, the more RNA they had, while the DNA template is constant. To achieve scaling, cells have to either increase the RNA production rate or increase RNA stability in bigger cells,” Dr Berry said.
While they identified hundreds of genes associated with the rate of RNA synthesis, systematic disruption of these genes seldom led to altered global RNA concentration, regardless of the size of the cell.
Feedback Mechanism
Mining of the extensive data generated led the researchers to proposed that RNA synthesis rates were regulated by the concentration of mRNA in the cell nucleus, where it is transcribed. The concentration of nuclear mRNA detected provides feedback to drive RNA metabolism to either boost or reduce the RNA content of the cell – by either making more mRNA copies or slowing the export from – or degradation of RNA in – the nucleus.
The deep dive into how cells change when mRNA synthesis or degradation are disrupted uncovered that RNA polymerase II – an enzyme that drives mRNA transcription – is instrumental in ramping up or dialling back RNA production when the cell senses a change in mRNA concentration.
“What we found is that this seems to be mediated through controlling the activity and ultimately, the abundance of the polymerase itself,” said Dr Berry.
“This paper brings together, for the first time, two disparate research areas: cell size-scaling and RNA metabolism. We see RNA buffering and cell-size scaling of transcription as two sides of the same coin. Hopefully this work builds a bridge between these two communities.”
Dr Berry and Prof Pelkmans provide further insight into the fundamental problem of how cells coordinate mRNA abundance with cell size to achieve mRNA concentration homeostasis in a recent review currently in press in Trends in Cell Biology.
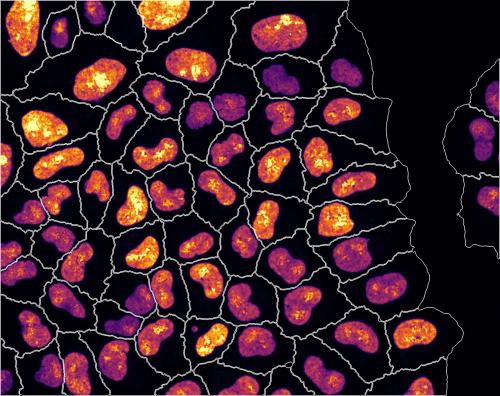
Metabolic labelling with fluorescent dye indicates rate of RNA production. Image analysis measured the size, growth phase and rate of RNA production of every single cell