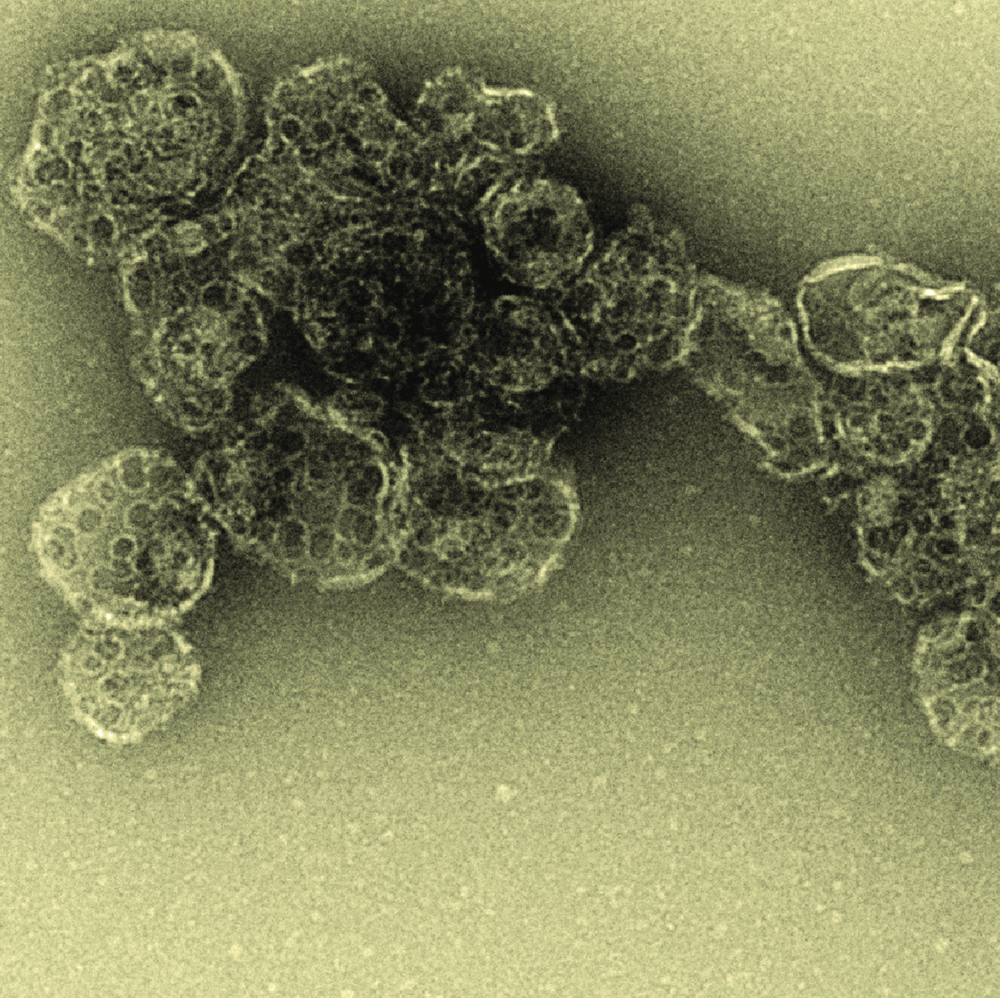
To combat viruses, host cells have developed weapons to sabotage the delivery of viral genetic code into their nuclei. There are specialised proteins that either prematurely crack open the protective viral capsid encasing the virus—exposing its genetic material to degradation—or lock the DNA in so it cannot enter the nucleus.
Till Böcking wants to understand how these and other molecular machines work.
By reconstituting these protein interactions—and watching them work using single molecule imaging—Till can see just what is required for the machines to run. He also hopes his findings will help in drug design to improve these machines and fight disease.
We develop fluorescence imaging approaches to visualise the dynamics of self-assembly processes at the single molecule level. The advantage of single molecule measurements is that they can resolve the kinetics of processes without the need for synchronisation and permit the detection of short-lived intermediates in the reaction pathways that are otherwise averaged out in traditional ensemble measurements.
Our work draws on approaches from the physical sciences with development of microfluidic imaging devices, surface chemistry approaches and development of automated image analysis software.
Please see our current projects below.
Our research focuses on elucidating the mechanisms of molecular machines in cellular assembly and disassembly processes using a combination of biochemical and biophysical approaches. In particular we develop fluorescence imaging approaches to visualise the dynamics of these processes at the single-molecule level. The advantage of single molecule measurements is that they can resolve the kinetics of processes without the need for synchronisation and permit the detection of short-lived intermediates in the reaction pathways that are otherwise averaged out in classical ensemble measurements. Our work draws on approaches from the physical sciences with development of microfluidic imaging devices, surface chemistry approaches and development of automated image analysis software.
Virus-host cell interactions
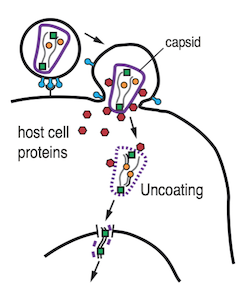
HIV infection depends on the safe passage of the viral genome to the nucleus. Strategies to manipulate viral gene delivery have two clinical applications: first for development of new HIV therapies and second for improving lentiviral vectors for gene therapy. A promising target for controlling viral gene delivery is HIV uncoating, a critical step in the life cycle that releases the viral genome from the viral capsid. Our aim is to identify host cell proteins that interact with the HIV capsid and elucidate their roles in HIV uncoating. The HIV-1 capsid is a protein shell protecting the viral genome after host cell entry and undergoes a highly regulated disassembly process termed “uncoating”. The virus has evolved to exploit a network of host cofactors that interact with the incoming capsid core to regulate reverse transcription, nuclear import and integration of viral cDNA into preferred regions of the host chromosome. Conversely the cell has evolved restriction factors that recognise the capsid lattice and block infection. Spatiotemporal control of uncoating plays a central role in these competing processes: Host cofactors function by controlling capsid stability to evade detection by innate immune sensors while restriction factors lead to premature uncoating (or prevent uncoating entirely), resulting in loss of infectivity and degradation of the virus. Despite the central role of the capsid in the viral life cycle, only few host capsid-binding proteins have been identified and their precise roles in uncoating are not well understood. We have developed a highly innovative single-molecule uncoating assay to visualise binding of host proteins and their effect on capsid disassembly in vitro. The assay allows us to pinpoint the time of capsid opening. We have observed that the first defects appear soon after release of the capsid from the viral envelope followed by lattice disassembly. Small molecule inhibitors known to destabilise the viral capsid lead to a pronounced acceleration of the capsid opening time. We aim to dissect the competing protein interaction networks that assemble on the capsid lattice as part of the arms race between host and virus. This project is a close collaboration with the team led by A/Prof Stuart Turville (Kirby Institute, UNSW) such that we can validate predictions from our in vitro studies of host cell factor competition in cellular models of HIV infection.
Scientia PhD project available: http://www.2025.unsw.edu.au/apply/scientia-phd-scholarships/molecular-arms-race-between-host-cells-and-hiv
Assembly pathways of the actin cytoskeleton
Actin filaments play a critical role as part of the cytoskeleton, where they are involved in most cellular processes including determination of cell shape, cell migration, cell division, membrane function and intracellular transport. This enormous functional specialisation is associated with differences in filament organisation, dynamics and interaction with actin-binding proteins. We aim to elucidate the fundamental molecular mechanisms that drive the self-assembly of functionally distinct actin filaments.
Scientia PhD scholarship available: http://www.2025.unsw.edu.au/apply/scientia-phd-scholarships/mathematical-biophysics-single-molecule-fluorescence-and-protein-protein

Molecular chaperones are molecular motors that drive a variety of intracellular assembly and disassembly processes to catalyse protein folding, prevent aggregation and facilitate the recycling of the components of macromolecular complexes. These functions are vital to cellular homeostasis and chaperone malfunction underlies a range of neurodegenerative diseases and cancers. However, the molecular mechanisms of these processes and how they can be targeted pharmacologically are not well understood. Capitalising on our recent single-molecule imaging approach to visualise chaperone dynamics (Figure1; Nat Struct Mol Biol 2011, 18(3), 295-301) we now aim extend our mechanistic studies of role of Hsp70 family chaperones in its diverse cellular functions.
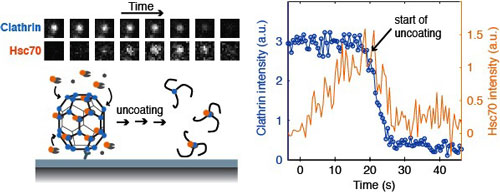
Figure 1: Fluorescence imaging approach to follow in real time the chaperone-mediated disassembly of the protein coat surrounding endocytic clathrin coated vesicles.Left: Selected snapshots from a time series (top) showing the fluorescence from clathrin and the chaperone Hsc70 in a coat visualised using the single-particle uncoating assay (shown schematically at the bottom). Right: The plot shows intensity traces of the clathrin (blue) and Hsc70 (orange) signals of a single coat during the uncoating reaction.
A long-standing interest is focused on controlling the properties of surfaces and materials at the molecular level using self-assembly chemistries. Our aim is to develop tools that enable us to investigate cellular processes quantitatively. Surface chemistry and nanotechnology also play in important role in devising strategies to capture and immobilise individual molecules or molecular assemblies for single molecule observation.
Group Leader
 |
Till Böcking+61 2 9385 1179
Till studied biochemistry at the University of Bonn in Germany before moving to Sydney for his PhD studies in biophysics followed by a short post-doc in chemistry. In 2006 he joined the group of Tomas Kirchhausen at Harvard Medical School in Boston to apply his background in the physical sciences to study endocytosis. Till was lured back to Sydney in 2011 where is now setting up the Molecular Machines group at UNSW. |
Postdoctoral Fellows |
|
 |
Dr Miro Jancom.janco@unsw.edu.au Miro received his Bachelor’s degree in Environmental Ecology at the Faculty of Natural Sciences, University of Matej Bel, Slovakia. Shortly after moving to England he was awarded the Undergraduate certificate with Distinction in Biology at University of Kent followed by Master’s degree in Biochemistry, both at University of Kent. During his Master’s studies he was investigating the effect of CO2 on the quorum sensing regulated virulence determinants in an opportunistic human pathogen Pseudomonas aeruginosaPAO 1. Miro then joined the Geeves lab at the University of Kent where he completed his PhD in Biochemistry. His PhD studies were focused on the effect of skeletal α-tropomyosin carrying cardiomyopathy mutations on the regulation of cardiac muscle contraction. Here in the Boecking group at UNSW he is aiming to resolve the mechanism of tropomyosin assembly on actin filaments and the processes involved in the selection of various tropomyosin isoforms coating actin filaments using a single molecule microscopy combined with microfluidics. |
 |
Dr Vaibhav Shah+61 2 9385 8639 Vaibhav obtained his Bachelors and Masters in Pharmacy from India. Thereafter, he completed his Ph.D. in Cell Biology/Signal Transduction at Ohio State University with a focus on understanding the cellular signaling pathways leading to activation of brain microglial cells. While studying cellular signaling in immune cells, he developed a keen interest in infectious diseases, and therefore joined Chris Aiken’s laboratory at Vanderbilt University to study mechanisms of HIV-1 uncoating. In 2013, he joined UNSW with an aim to understand molecular mechanisms and effects of host factors on HIV-1 uncoating using imaging techniques. |
|
|
James WalshJames completed a Bachelor of science in Physics/Mathematics at UNSW in 2009 before obtaining first class Honours in Biophysics in 2010. He completed his PhD in 2015 under the supervision of Paul Curmi where his work concentrated on the mechanisms of division plane positioning in Bacteria and Archae, focusing on “Turing Pattern-like” mechanisms to sense the geometry of the cell. In 2016, James joined the Molecular Machines group where he has been focusing on the studying the kinetics of pore formation by cytolytic proteins, as well as the development of image acquisition automation and analysis software. |
PhD Candidates |
|
 |
Ilina BarejaI received a Bachelor's degree in Biochemistry from the University of Delhi, India and then went on to pursue a master's degree in Biotechnology from the Indian Institute of Technology, Bombay, India. For my dissertation I worked to look at the effect of a novel centrosomal protein, FOR20, on microtubule assembly in vitro. My PhD project focuses on studying the interactions between actin and different tropomyosin isoforms using an in vitro reconstitution system, and to help elucidate how different isoforms may compete for this binding, as well as the effect of certain anti-cancer drugs on the actin filament assembly. |
 |
Chantal Marquez BadillaChantal received her Bachelor’s degree in Molecular Biotechnology Engineering at University of Chile. To obtain the professional degree she worked with Hantaviruses, specifically characterising the role of the transmembrane domain of the Gc fusion protein. After moving to Sydney she joined the Molecular Machines group for a short internship, were she stayed to do her PhD. Chantal’s project focuses on understanding the molecular mechanism of HIV uncoating, for which she is developing a new assay to detect capsid opening and to visualise the interaction of host cell factors with the viral core. |
 |
Derrick LauDerrick completed a combined Bachelors in Commerce and Science at University of New South Wales in 2014. He further pursued his passion for research into infectious diseases and molecular biology during his Honours project with Till Böcking. Derrick shown that it is possible to assemble fluorescently labelled capsid lattices for fluorescent microscopy to develop new tools for studying protein-capsid interactions that occur during the early phase of the viral life cycle. As time flew, this project transformed into a PhD project. Using self-assembled capsid lattices Derrick hopes to understand how HIV utilises various host cell factors to mask itself from detection in the host cell and to control capsid uncoating. |
 |
Jeffrey Stear |
Philip Nicovich
Physicist
University of New South Wales
http://www.bmif.unsw.edu.au/aboutus.htm
Paul Curmi
Physicist
University of New South Wales
https://research.unsw.edu.au/people/professor-paul-marie-gerard-curmi
Danny Hatters
University of Melbourne
http://www.biochemistry.unimelb.edu.au/research/res_hatters.html
Adelle Coster
Biomathematician
University of New South Wales
https://research.unsw.edu.au/people/dr-adelle-coster
For more information please contact:
E: Till.Boecking@unsw.edu.au